Develop new quantitative modeling methods and advanced computational approaches to advance understanding of biological systems and improve human health.
About the Program
It is widely anticipated that quantitative modeling, advanced computing and data science will transform the biomedical research enterprise and practice of medicine in the coming decades as fundamentally as biochemistry and molecular biology transformed it during the past century. With leading researchers from seven institutions, the Quantitative & Computational Biosciences Graduate Program brings together the resources of the Texas Medical Center -- the world's largest complex of biomedical research institutions and hospitals, Rice University and neighboring institutions -- to discover new biomedical knowledge and improve human health.
Faculty
Our full-time faculty includes basic and clinical scientists from seven institutions -- Baylor College of Medicine, Methodist Research Institute, Rice University, University of Houston, University of Texas Health Science Center, University of Texas Medical Branch at Galveston and University of Texas MD Anderson Cancer Center.
Where Will Your Ph.D. Take You?
From day one, we encourage you to think deeply about your career choices. Wherever your ambition leads, you will receive the support you need to follow a path well worn by our alumni who have built successful careers across diverse endeavors.
QCB News

PolyA-miner accurately assesses the effect of alternative polyadenylation on gene expression
Researchers with an interest in unraveling gene regulation in human health and disease are expanding their horizons by closely looking at alternative polyadenylation (APA), an under-charted mechanism that regulates gene expression. The interest in APA has resulted in the development of several 3′ sequencing (3′Seq) techniques that allow for precise identification on APA sites on RNA strands. But what researchers are missing is a robust computational tool that is specifically designed to analyze the wealth of 3′Seq data that has been generated. Baylor researchers developed a tool for this they call PolyA-miner.
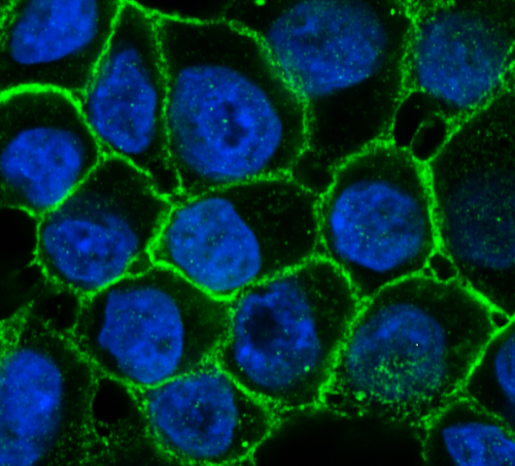
AKAP8 and the delicate balance between normal and metastatic states
A protein naturally produced in the body has been found to suppress breast cancer metastasis in animal models of human tumors. In this study, the same team at BCM that published a pioneering study that brought alternative splicing to the field of cancer research, investigated how alternative splicing contributes to cancer metastasis by looking for proteins that regulate alternative splicing events linked to metastasis.
An unprecedented look at endometrial cancer
Which therapy will benefit this particular patient the most? BCM researchers and colleagues offer insights that might help physicians better answer this question for endometrial cancer, commonly known as uterine cancer.
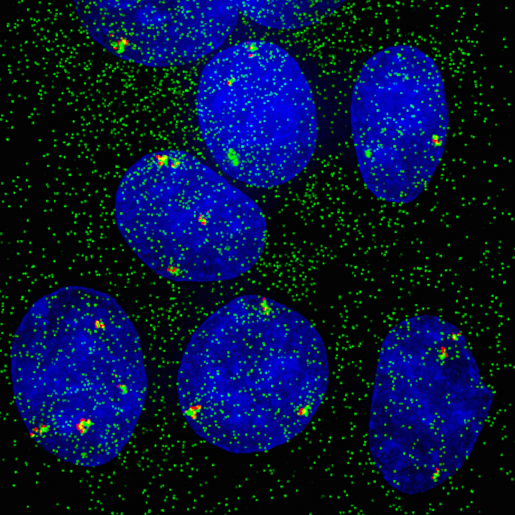
Not in sync: single cells and genes show unanticipated response to estrogen
When experimental findings are not compatible with the textbook view, scientists know they have quite a challenge in their hands. BCM researchers and their colleagues faced this situation when they were investigating how cells modulate their responses to estrogen at single-cell and -gene level. In the current study, researchers worked with human breast cancer cell lines grown in the lab. Using both molecular and imaging analyses, they determined, at single-cell and -allele levels, the expression of two well-characterized genes whose activity is regulated by estrogen.
A closer look at severe malnutrition
Severe acute childhood malnutrition presents in two clinically distinct forms: edematous severe acute malnutrition (ESAM) and non-edematous SAM (NESAM). Children with ESAM tend to show body swelling and extensive dysfunction of multiple organs, including liver, blood cells and the gut, as well as skin and hair abnormalities. NESAM, on the other hand, typically presents with weight loss and wasting. BCM researchers and colleagues looked at these conditions from a molecular perspective. Specifically, they investigated whether DNA methylation differed between ESAM and NESAM. Their findings suggest novel opportunities for improving these conditions in the future.
The herpes virus dilemma in Alzheimer’s disease
There is currently a debate about whether herpes virus infections play a role in Alzheimer’s disease and BCM researchers are actively contributing to the discussion. They looked into the proposed link between herpes virus infections and Alzheimer’s disease and found unexpected results.

From the Labs
Keep up-to-date on all the research news from Baylor College of Medicine. Subscribe to our From the Labs blog.
Graduate School of Biomedical Sciences
The Quantitative and Computational Biosciences Graduate Program is part of the Baylor College of Medicine Graduate School of Biomedical Sciences. Visit the GSBS website for information about our curriculum and admissions process as well as to find resources and services designed to support your success throughout graduate school and your future career.
Stipends and Benefits
At BCM, we are focused on you and your training. If your vision for the future includes teaching, you may choose to gain experience as a teaching assistant. If you do not want to teach, you have the freedom to focus exclusively on your education and research as well as to work with your mentor to take advantage of other BCM resources that match your interests.











