Copy-scAT: A new tool that uses single-cell ATAC-seq data to infer copy number variation
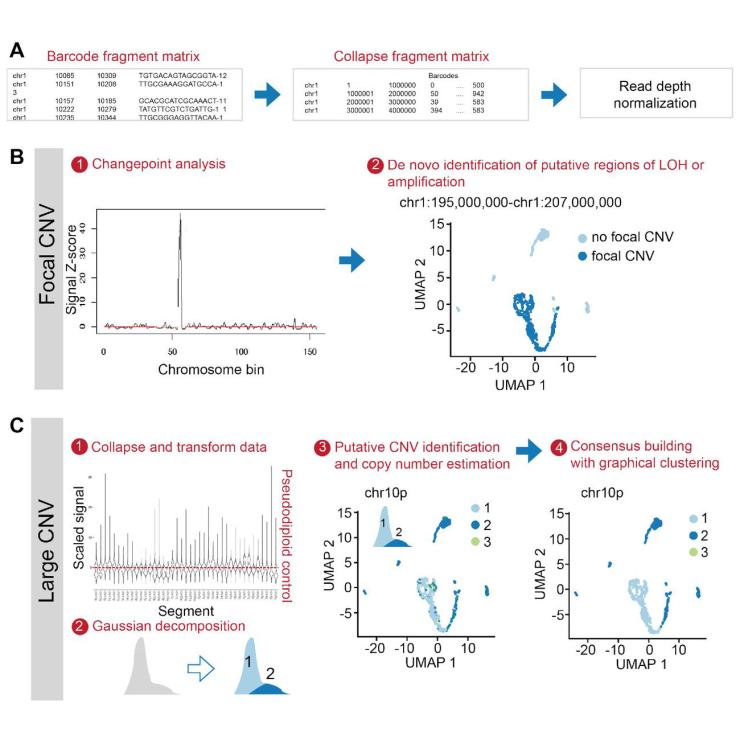
We have created a new R package called Copy-scAT (copy number inference using single-cell ATAC-seq data) to infer copy number genetic alterations using scATAC-seq data.
When performing scATAC using clinical tumour samples – especially solid malignancies – libraries will be generated for both malignant and non-malignant cells (eg tumour-infiltrating cells and other cells normally found in the tumour microenvironment). Inability to distinguish malignant and non-malignant cells can confound downstream analyses.
Copy-scAT will be an important tool for scientists who are interested in studying epigenetic profiles at the single-cell level in clinical cancer samples.
You can read more on Copy-scAT in our preprint. If you want to try Copy-scAT, you can find instructions and a tutorial on GitHub.
Exploring Candidate Cancer-Related Germline Variants
Traditionally, two main types of mutations are thought to lead to cancer:
(a) Germline mutations inherited by at least one parent often cause familial cancer syndromes
(b) Other genetic lesions, called somatic mutations, arise specifically in tumour cells and contribute to the growth and spread of the malignancy
One of our recent publications, however, showed that pediatric high-grade glioma patients can also have germline mutations that are not inherited, and seem to arise specifically in the patient.
Because of this strong emerging need to identify germline variants linked to pediatric cancer, we explored variables that could confound the identification of cancer-predisposing mutations.
Work by a very talented undergraduate student in our lab, Anna Bobyn, has showcased the need to consider ancestry when looking for candidate cancer-associated germline variants. For instance, we found that deletions upstream of a gene called NEGR1 are found at different frequencies in different ethnic groups.
Anna Bobyn’s paper suggests a pipeline to account for ancestry in the identification of germline variants associated with cancer and was published on May 6, 2020, in BMC Medical Genetics.
3D Genome Architecture of Cancer Stem Cells
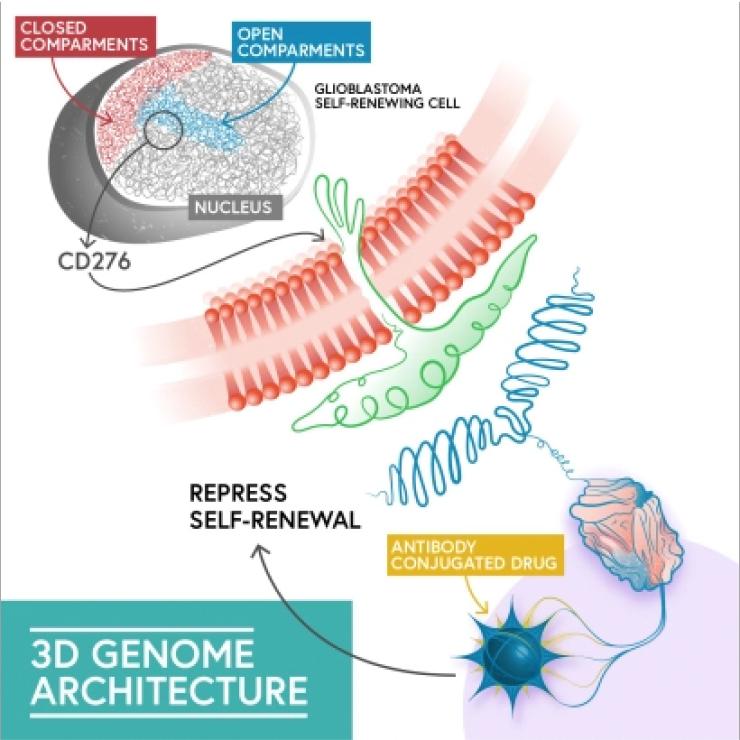
One of our projects on 3D genome architecture was recently published in Genome Research. This project was led by Michael Johnston, a postdoc in our lab.
Michael generated very high-resolution contact maps of cancer stem cells using in situ Hi-C. We were able to define compartments and domains and to visualize individual chromatin loops linking regulatory regions of the genome with the target gene.
By integrating 3D genome maps and other chromatin and transcriptional datasets that we generated, we were able to dissect precise mechanisms of regulation of gene expression.
Among the main findings, we showed that stem cell genes have a tendency to be in contact with multiple super-enhancers concurrently. These arrangements might ensure sustained expression of these key genes.
We also found that the 3D genome of cancer stem cells is shaped in such a way as to enable the expression of immune-related genes. Among these genes, we focused on CD276, which has a putative role in suppressing T cell function. We found that, unexpectedly, CD276 has a role in maintaining some key stemness properties in GBM cells. We also deployed an antibody-drug conjugate and showed that targeting CD276 may be an effective strategy to kill cancer stem cells in GBM.
Evolutionary Trajectories in Pediatric GBM

Our manuscript on genomic evolution in pediatric GBM was accepted for publication in Cancer Research and is now available on the journal website.
This project was led by Mary Hoffman, an MSc student in my lab. Her work showed that pediatric GBM harbors many genetic subclones – basically, each tumor contains many mini-tumors, each with its own specific genomes.
In addition, our results indicate that pediatric GBM may be driven by populations of slow-cycling cancer stem cells. Because they do not divide very often, these cancer stem cells may be able to escape chemotherapy and radiotherapy during treatment and may be responsible to re-grow the tumor at relapse.
Overall, Mary’s work showed that pediatric GBM is a complex ecosystem populated by cells with different genomes and functional properties, clearly challenging long-held views that postulated childhood cancers are relatively homogeneous malignancies.
Future efforts in our lab will be aimed at identifying new therapeutic opportunities that specifically target the slow-cycling cancer stem cell populations.








