Deep sequencing has revealed thousands of eukaryotic protein-coding genes that defy the central dogma, producing circular noncoding RNAs rather than linear messenger RNAs. For some genes, the abundance of the circular RNA exceeds that of the associated linear mRNA by a factor of 10, raising the interesting possibility that the function of some protein-coding genes may actually be to produce circular noncoding RNAs, not proteins. These circular RNAs are generated when the pre-mRNA splicing machinery “backsplices” and joins a splice donor to an upstream splice acceptor (Figure 1). Once produced, it is largely unclear what circular RNAs do, although two are known to efficiently modulate the activity of microRNAs.
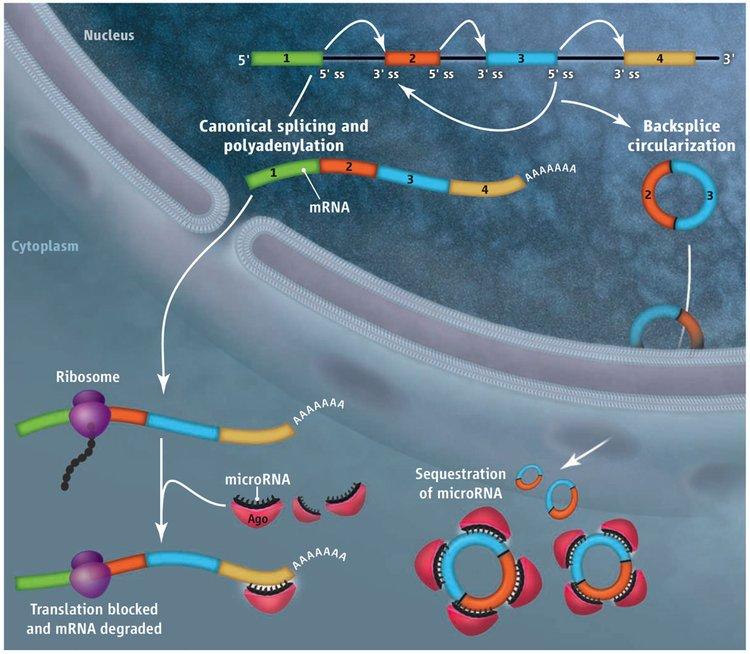
With the exception of the first and last exons of genes, every other exon in the genome has splicing signals at its 5’ and 3’ ends and theoretically can circularize. However, every exon does not circularize, and, in some cases, multiple exons are present in a circular RNA. We showed that repetitive elements, e.g. SINE elements, in the flanking introns are critical determinants of whether the intervening exon(s) circularize. When repeat sequences from the flanking introns base pair to one another, the splice sites are brought into close proximity and backsplicing occurs. Although mechanistically simple, this step occurs in a highly selective manner, as the sequence of the repeats can drastically alter the efficiency of circular RNA production.
How then is the ratio of linear to circular RNA controlled or modulated? Using RNAi screening in Drosophila cells, we identified many hnRNPs, SR proteins, core spliceosome, and transcription termination factors that control the outputs of reporter and endogenous genes. Surprisingly, when spliceosome components were depleted or inhibited pharmacologically, the steady-state levels of circular RNAs increased while expression of their associated linear mRNAs concomitantly decreased. We propose that this is because cross-exon interactions are not easily replaced with cross-intron interactions, thereby causing spliceosomes to preferentially assemble across an exon and generate a circular RNA. Upon inhibiting RNA polymerase II termination, circular RNA levels were similarly increased due to readthrough transcripts extending into downstream genes and being subjected to backsplicing. In total, these results indicate that inhibition or slowing of canonical pre-mRNA processing events shifts the steady-state output of protein-coding genes towards circular RNAs, which likely helps explain why and how circular RNAs show tissue-specific expression profiles.
Most mature circular RNAs accumulate in the cytoplasm, and we revealed the first insights into how their nuclear export is controlled in a length-dependent manner. We further developed improved methods for circular RNA identification and purification using RNase R by identifying a way to better remove linear RNAs containing G-quadruplexes or structured 3’ ends.
By revealing the fundamental mechanisms by which circular RNAs are generated, we have developed plasmid and viral-based methods for ectopically expressing circular RNAs. Almost any sequence can now be efficiently circularized in eukaryotic cells, which has enabled us (and others) to begin to ask how circular RNAs function. More than 150 labs have requested our circular RNA expression plasmids, highlighting our wide impact on this growing field.
We are continuing to elucidate the mechanism by which circular RNAs are produced. In particular, we are interested in determining how cellular cues can alter the ratio of linear mRNA to circular RNA for a given gene. We are also focused on identifying biological functions for circular RNAs, thereby revealing novel insights into how circular RNAs fit into the regulatory landscape of the cell.
Key Publications
-
Xiao, M.S. and Wilusz, J.E. (2019) An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids Res 47: 8755-8769.
-
Huang, C., Liang, D., Tatomer, D.C., and Wilusz, J.E. (2018) A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32: 639-644.
-
Liang, D., Tatomer, D.C., Luo, Z., Wu, H., Yang, L., Chen, L.L., Cherry, S., and Wilusz, J.E. (2017) The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell 68: 940-954.
-
Tatomer, D.C., Liang, D., and Wilusz, J.E. (2017) Inducible expression of eukaryotic circular RNAs from plasmids. Methods Mol Biol 1648: 143-154.
-
Chen, Y.G., Kim, M.V., Chen, X., Batista, P.J., Aoyama, S., Wilusz, J.E., Iwasaki, A., and Chang, H.Y. (2017) Sensing self and foreign circular RNAs by intron identity. Mol Cell 67: 228-238.
-
Kramer, M.C., Liang, D., Tatomer, D.C., Gold, B., March, Z.M., Cherry, S., and Wilusz, J.E. (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29: 2168-2182.
-
Liang, D. and Wilusz, J.E. (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233-2247.
-
Wilusz, J.E. and Sharp, P.A. (2013) A circuitous route to noncoding RNA. Science 340: 440-441.








