About This Lab
In the Herman Lab, we set out to exploit the speed and power of bacterial genetics to understand the interactions between transcription, core transcription factors, and genome maintenance.
Specifically, we use the most heavily studied model organism, Escherichia coli, as a platform for understanding how RNA polymerase associated factors influence the fidelity of transcription as well as transcriptional interactions with DNA repair machinery.
Our research has also piqued our interest in microbial horizontal gene transfer. In particular, we study the fundamental mechanics and principles that influence bacterial conjugation. Our goal is to use this knowledge to facilitate the use of conjugation as a synthetic biological tool.

Lab photo 2021 at the BCM fountain.

Lab photo from Phages and Bacteria meeting 2022.
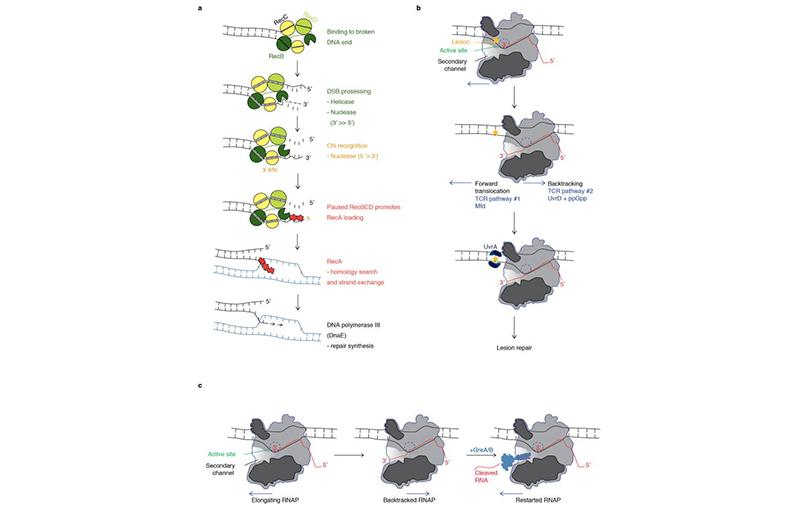
Genome Stability
It’s known that the genome stability of a cell is a major determinant of its fitness, as genomic strand breaks, re-arrangements, or single nucleotide mutations stand to compromise proper cell division, global transcriptional programs, and the expression of essential genes. It is also known that proper transcription of the same genome is a determinant of cellular fitness as mis-transcribed or partially transcribed mRNA’s lead to many defective proteins that are toxic to the cell. However, both of these processes, DNA replication and RNA transcription, occur on the same template DNA. As such, they are bound to conflict.
Our group and our collaborators have made major breakthroughs in understanding how secondary channel factors interact with DNA replication and DNA repair; to assist or conflict with the stability of the genome.

Transcription Infidelity
Much of modern molecular genetics has been concerned with the concept of error propagation in DNA. In basic research, we refer to these as mutations – our more clinically inclined friends: variants. Semantics aside, these mutations are the basis of long term cellular adaptation; they are the only event that can heritably change a cell’s phenotype and fitness – or are they?
Our lab aims to adjust this notion and make a case that errors in transcriptional RNA propagation are also able to influence cellular phenotypes, and subsequently, fitness.
Our lab has demonstrated that these RNA error-based “epimutations” do indeed result in heritable, phenotypic heterogeneity.
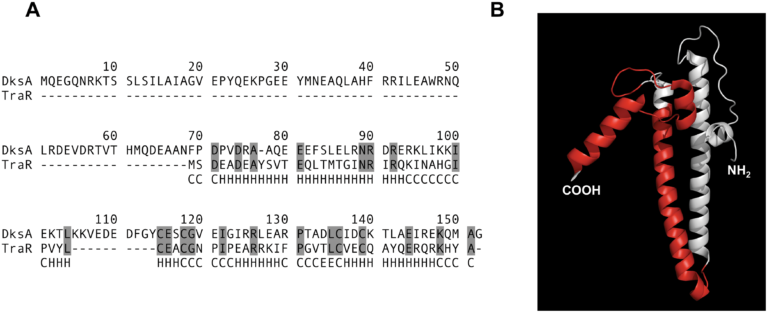
Horizontal Gene Transfer
Microbes frequently acquire genes from others in a process called horizontal gene transfer (HGT). Many processes can contribute to HGT, including phage-mediated transduction or natural transformation, however, we’ve become interested in a particular process called conjugation. Conjugation is mediated by an autonomous genetic unit that commonly comes in a plasmid or prophage-like form. This genetic unit encodes for a massive cell-envelope mounted secretion system called a Type 4 Secretion System (T4SS). This T4SS is responsible for binding a recipient cell and transferring its own genetic element to the new host.
We became interested in these conjugative systems because they have a tendency of interacting with core host processes. The first example of this came when a member of the lab discovered that the most heavily studied conjugative system, F, encoded a dksA homolog called traR. This protein seems to manipulate the transcriptional state of the host cell to favor productive conjugative transfer.
As we move forward, we hope to gain a better understanding of how these systems interface with their hosts and work to spread to broader and broader populations.
Contact
Stop by or email to set up a rotation or apply for an interview.
Faculty Appointments:
Genetics and Genomics (Primary)
Immunology and Microbiology
Development, Disease Models & Therapeutics
Cancer and Cell Biology
Lab Address: BCM Main Campus, Taub Building, T913/T907
Contact Email: herman@bcm.edu








