
Xia J, Chiu LY, Nehring RB, Bravo Núñez MA, Mei Q, Perez M, Zhai Y, Fitzgerald DM, Pribis JP, Wang Y, Hu CW, Powell RT, LaBonte SA, Jalali A, Matadamas Guzmán ML, Lentzsch AM, Szafran AT, Joshi MC, Richters M, Gibson JL, Frisch RL, Hastings PJ, Bates D, Queitsch C, Hilsenbeck SG, Coarfa C, Hu JC, Siegele DA, Scott KL, Liang H, Mancini MA, Herman C, Miller KM, Rosenberg SM. Bacteria-to-Human Protein Networks Reveal Origins of Endogenous DNA Damage. Cell. 2019 Jan 10;176(1-2):127-143.e24. doi: 10.1016/j.cell.2018.12.008. PMID: 30633903; PMCID: PMC6344048.

Sivaramakrishnan P, Sepúlveda LA, Halliday JA, Liu J, Núñez MAB, Golding I, Rosenberg SM, Herman C. The transcription fidelity factor GreA impedes DNA break repair. Nature. 2017 Oct 12;550(7675):214-218. doi: 10.1038/nature23907. Epub 2017 Oct 4. PMID: 28976965; PMCID: PMC5654330.

Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, Sowa JN, Sizovs A, Du G, Wang J, Herman C, Wang MC. Microbial Genetic Composition Tunes Host Longevity. Cell. 2017 Jun 15;169(7):1249-1262.e13. doi: 10.1016/j.cell.2017.05.036. Erratum in: Cell. 2018 May 3;173(4):1058. PMID: 28622510; PMCID: PMC5635830.
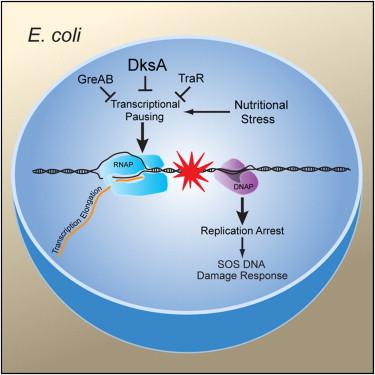
Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010 May 14;141(4):595-605. doi: 10.1016/j.cell.2010.03.036. PMID: 20478253; PMCID: PMC2919171.

Gordon AJ, Satory D, Halliday JA, Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013 Jun;9(6):e1003595. doi: 10.1371/journal.pgen.1003595. Epub 2013 Jun 27. PMID: 23825966; PMCID: PMC3694819.

Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2009 Jan;5(1):e1000345. doi: 10.1371/journal.pgen.1000345. Epub 2009 Jan 16. PMID: 19148274; PMCID: PMC2613031.

Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003 Mar;11(3):659-69. doi: 10.1016/s1097-2765(03)00068-6. PMID: 12667449.

Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D’Ari R, Bouloc P. Cell growth and lambda phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10861-5. doi: 10.1073/pnas.90.22.10861. PMID: 8248182; PMCID: PMC47878.
Pre Prints
Mindy Engevik, Lei Guo, Won-Suk Song, Christopher G Chronowski, Viktor Akhanov, Navish Bosquez, Amy C. Engevik, Numan Oezguen, Cristian Coarfa, Nagireddy Putluri, Andre Catic, Joseph A. Baur, Milton Finegold, Noah Shroyer, Jianhua Gu, Ronaldo P. Ferraris, Jennifer A Halliday, Christophe Herman, Simona Colla, Herman Dierick, Cholsoon Jang, Ergun Sahin. Telomere dysfunction impairs intestinal differentiation and predisposes to diet-induced colitis. bioRxiv 2022.07.14.499988; doi: https://doi.org/10.1101/2022.07.14.499988.
Yin Zhai, P.J. Minnick, John P. Pribis, Libertad Garcia-Villada, P.J. Hastings, Christophe Herman, Susan M. Rosenberg. ppGpp and RNA-Polymerase Backtracking Guide Antibiotic-Induced Mutable Gambler Cells. bioRxiv 2022.07.06.499071; doi: https://doi.org/10.1101/2022.07.06.499071.
Christophe Herman, Catherine Bradley, Alasdair Gordon, Chen Wang, Matthew Cooke, Brendan Kohrn, Scott Kennedy, Olivier Lichtarge, Shannon Ronca. RNA polymerase inaccuracy underlies SARS-CoV-2 variants and vaccine heterogeneity.
Priya Sivaramakrishnan, Catherine C. Bradley, Irina Artsimovitch, Katelin M. Hagstrom, Laura Deus Ramirez, Alice Xueyin Wen, Matthew Brandon Cooke, Maya Shaulsky, Christophe Herman, Jennifer A. Halliday. Modulation of RNA polymerase processivity affects double-strand break repair in the presence of a DNA end-binding protein. bioRxiv 2022.02.08.479637; doi: https://doi.org/10.1101/2022.02.08.479637.
All Publications
Neve IAA, Sowa JN, Lin CJ, Sivaramakrishnan P, Herman C, Ye Y, Han L, Wang MC. Escherichia coli Metabolite Profiling Leads to the Development of an RNA Interference Strain for Caenorhabditis elegans. G3 (Bethesda). 2020 Jan 7;10(1):189-198. doi: 10.1534/g3.119.400741. PMID: 31712257; PMCID: PMC6945014.
Bradley CC, Gordon AJE, Halliday JA, Herman C. Transcription fidelity: New paradigms in epigenetic inheritance, genome instability and disease. DNA Repair (Amst). 2019 Sep;81:102652. doi: 10.1016/j.dnarep.2019.102652. Epub 2019 Jul 8. PMID: 31326363; PMCID: PMC6764924.
Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald DM, Bos J, Austin RH, Herman C, Bates D, Hadany L, Hastings PJ, Rosenberg SM. Gamblers: An Antibiotic-Induced Evolvable Cell Subpopulation Differentiated by Reactive-Oxygen-Induced General Stress Response. Mol Cell. 2019 May 16;74(4):785-800.e7. doi: 10.1016/j.molcel.2019.02.037. Epub 2019 Apr 1. PMID: 30948267; PMCID: PMC6553487.
Xia J, Chiu LY, Nehring RB, Bravo Núñez MA, Mei Q, Perez M, Zhai Y, Fitzgerald DM, Pribis JP, Wang Y, Hu CW, Powell RT, LaBonte SA, Jalali A, Matadamas Guzmán ML, Lentzsch AM, Szafran AT, Joshi MC, Richters M, Gibson JL, Frisch RL, Hastings PJ, Bates D, Queitsch C, Hilsenbeck SG, Coarfa C, Hu JC, Siegele DA, Scott KL, Liang H, Mancini MA, Herman C, Miller KM, Rosenberg SM. Bacteria-to-Human Protein Networks Reveal Origins of Endogenous DNA Damage. Cell. 2019 Jan 10;176(1-2):127-143.e24. doi: 10.1016/j.cell.2018.12.008. PMID: 30633903; PMCID: PMC6344048.
Kotlajich MV, Xia J, Zhai Y, Lin HY, Bradley CC, Shen X, Mei Q, Wang AZ, Lynn EJ, Shee C, Chen LT, Li L, Miller KM, Herman C, Hastings PJ, Rosenberg SM. Fluorescent fusions of the N protein of phage Mu label DNA damage in living cells. DNA Repair (Amst). 2018 Dec;72:86-92. doi: 10.1016/j.dnarep.2018.09.005. Epub 2018 Sep 14. PMID: 30268364; PMCID: PMC6287932.
Gordon AJE, Sivaramakrishnan P, Halliday JA, Herman C. Transcription infidelity and genome integrity: the parallax view. Transcription. 2018;9(5):315-320. doi: 10.1080/21541264.2018.1491251. Epub 2018 Aug 10. PMID: 29929421; PMCID: PMC6150618.
Sivaramakrishnan P, Gordon AJE, Halliday JA, Herman C. How Acts of Infidelity Promote DNA Break Repair: Collision and Collusion Between DNA Repair and Transcription. Bioessays. 2018 Oct;40(10):e1800045. doi: 10.1002/bies.201800045. Epub 2018 Aug 9. PMID: 30091472; PMCID: PMC6334755.
Swings T, Marciano DC, Atri B, Bosserman RE, Wang C, Leysen M, Bonte C, Schalck T, Furey I, Van den Bergh B, Verstraeten N, Christie PJ, Herman C, Lichtarge O, Michiels J. CRISPR-FRT targets shared sites in a knock-out collection for off-the-shelf genome editing. Nat Commun. 2018 Jun 8;9(1):2231. doi: 10.1038/s41467-018-04651-5. PMID: 29884781; PMCID: PMC5993718.
Girard ME, Gopalkrishnan S, Grace ED, Halliday JA, Gourse RL, Herman C. DksA and ppGpp Regulate the σS Stress Response by Activating Promoters for the Small RNA DsrA and the Anti-Adapter Protein IraP. J Bacteriol. 2017 Dec 20;200(2):e00463-17. doi: 10.1128/JB.00463-17. PMID: 29061665; PMCID: PMC5738727.
Sivaramakrishnan P, Sepúlveda LA, Halliday JA, Liu J, Núñez MAB, Golding I, Rosenberg SM, Herman C. The transcription fidelity factor GreA impedes DNA break repair. Nature. 2017 Oct 12;550(7675):214-218. doi: 10.1038/nature23907. Epub 2017 Oct 4. PMID: 28976965; PMCID: PMC5654330.
Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, Sowa JN, Sizovs A, Du G, Wang J, Herman C, Wang MC. Microbial Genetic Composition Tunes Host Longevity. Cell. 2017 Jun 15;169(7):1249-1262.e13. doi: 10.1016/j.cell.2017.05.036. Erratum in: Cell. 2018 May 3;173(4):1058. PMID: 28622510; PMCID: PMC5635830.
Xia J, Chen LT, Mei Q, Ma CH, Halliday JA, Lin HY, Magnan D, Pribis JP, Fitzgerald DM, Hamilton HM, Richters M, Nehring RB, Shen X, Li L, Bates D, Hastings PJ, Herman C, Jayaram M, Rosenberg SM. Holliday junction trap shows how cells use recombination and a junction-guardian role of RecQ helicase. Sci Adv. 2016 Nov 18;2(11):e1601605. doi: 10.1126/sciadv.1601605. PMID: 28090586; PMCID: PMC5222578.
Marciano DC, Lua RC, Herman C, Lichtarge O. Cooperativity of Negative Autoregulation Confers Increased Mutational Robustness. Phys Rev Lett. 2016 Jun 24;116(25):258104. doi: 10.1103/PhysRevLett.116.258104. Epub 2016 Jun 22. PMID: 27391757; PMCID: PMC5152588.
Satory D, Gordon AJ, Wang M, Halliday JA, Golding I, Herman C. DksA involvement in transcription fidelity buffers stochastic epigenetic change. Nucleic Acids Res. 2015 Dec 2;43(21):10190-9. doi: 10.1093/nar/gkv839. Epub 2015 Aug 24. PMID: 26304546; PMCID: PMC4666387.
Gordon AJ, Satory D, Halliday JA, Herman C. Lost in transcription: transient errors in information transfer. Curr Opin Microbiol. 2015 Apr;24:80-7. doi: 10.1016/j.mib.2015.01.010. Epub 2015 Jan 28. PMID: 25637723; PMCID: PMC4380820.
Grace ED, Gopalkrishnan S, Girard ME, Blankschien MD, Ross W, Gourse RL, Herman C. Activation of the σE-dependent stress pathway by conjugative TraR may anticipate conjugational stress. J Bacteriol. 2015 Mar;197(5):924-31. doi: 10.1128/JB.02279-14. Epub 2014 Dec 22. PMID: 25535270; PMCID: PMC4325106.
Gordon AJ, Satory D, Wang M, Halliday JA, Golding I, Herman C. Removal of 8-oxo-GTP by MutT hydrolase is not a major contributor to transcriptional fidelity. Nucleic Acids Res. 2014 Oct 29;42(19):12015-26. doi: 10.1093/nar/gku912. Epub 2014 Oct 7. PMID: 25294823; PMCID: PMC4231768.
Marciano DC, Lua RC, Katsonis P, Amin SR, Herman C, Lichtarge O. Negative feedback in genetic circuits confers evolutionary resilience and capacitance. Cell Rep. 2014 Jun 26;7(6):1789-95. doi: 10.1016/j.celrep.2014.05.018. Epub 2014 Jun 5. PMID: 24910431; PMCID: PMC4103627.
Bednarz M, Halliday JA, Herman C, Golding I. Revisiting bistability in the lysis/lysogeny circuit of bacteriophage lambda. PLoS One. 2014 Jun 25;9(6):e100876. doi: 10.1371/journal.pone.0100876. PMID: 24963924; PMCID: PMC4070997.
Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, Landick R, Wang JD. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell. 2014 Mar 6;53(5):766-78. doi: 10.1016/j.molcel.2014.02.005. PMID: 24606919; PMCID: PMC4023959.
Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu LY, Magnan D, Halliday JA, Frisch RL, Gibson JL, Nehring RB, Do HG, Hernandez M, Li L, Herman C, Hastings PJ, Bates D, Harris RS, Miller KM, Rosenberg SM. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife. 2013 Oct 29;2:e01222. doi: 10.7554/eLife.01222. PMID: 24171103; PMCID: PMC3809393.
Satory D, Halliday JA, Sivaramakrishnan P, Lua RC, Herman C. Characterization of a novel RNA polymerase mutant that alters DksA activity. J Bacteriol. 2013 Sep;195(18):4187-94. doi: 10.1128/JB.00382-13. Epub 2013 Jul 12. PMID: 23852871; PMCID: PMC3754754.
Gordon AJ, Satory D, Halliday JA, Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013 Jun;9(6):e1003595. doi: 10.1371/journal.pgen.1003595. Epub 2013 Jun 27. PMID: 23825966; PMCID: PMC3694819.
Adikesavan AK, Katsonis P, Marciano DC, Lua R, Herman C, Lichtarge O. Separation of recombination and SOS response in Escherichia coli RecA suggests LexA interaction sites. PLoS Genet. 2011 Sep;7(9):e1002244. doi: 10.1371/journal.pgen.1002244. Epub 2011 Sep 1. PMID: 21912525; PMCID: PMC3164682.
Satory D, Gordon AJ, Halliday JA, Herman C. Epigenetic switches: can infidelity govern fate in microbes? Curr Opin Microbiol. 2011 Apr;14(2):212-7. doi: 10.1016/j.mib.2010.12.004. PMID: 21496764.
Gibson JL, Lombardo MJ, Thornton PC, Hu KH, Galhardo RS, Beadle B, Habib A, Magner DB, Frost LS, Herman C, Hastings PJ, Rosenberg SM. The sigma(E) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol Microbiol. 2010 Jul;77(2):415-30. doi: 10.1111/j.1365-2958.2010.07213.x. Epub 2010 May 19. PMID: 20497332; PMCID: PMC2909356.
Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010 May 14;141(4):595-605. doi: 10.1016/j.cell.2010.03.036. PMID: 20478253; PMCID: PMC2919171.
Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009 Jul 10;284(28):18674-84. doi: 10.1074/jbc.M900783200. Epub 2009 Apr 21. PMID: 19383601; PMCID: PMC2707231.
Blankschien MD, Lee JH, Grace ED, Lennon CW, Halliday JA, Ross W, Gourse RL, Herman C. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J. 2009 Jun 17;28(12):1720-31. doi: 10.1038/emboj.2009.126. Epub 2009 May 7. PMID: 19424178; PMCID: PMC2699356.
Gordon AJ, Halliday JA, Blankschien MD, Burns PA, Yatagai F, Herman C. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009 Feb 24;7(2):e44. doi: 10.1371/journal.pbio.1000044. PMID: 19243224; PMCID: PMC2652393.
Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2009 Jan;5(1):e1000345. doi: 10.1371/journal.pgen.1000345. Epub 2009 Jan 16. PMID: 19148274; PMCID: PMC2613031.
Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006 Jul 1;20(13):1776-89. doi: 10.1101/gad.1428206. PMID: 16818608; PMCID: PMC1522074.
Guisbert E, Herman C, Lu CZ, Gross CA. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004 Nov 15;18(22):2812-21. doi: 10.1101/gad.1219204. PMID: 15545634; PMCID: PMC528900.
Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004 Nov 1;18(21):2686-97. doi: 10.1101/gad.1238604. PMID: 15520285; PMCID: PMC525548.
Kobiler O, Oppenheim AB, Herman C. Recruitment of host ATP-dependent proteases by bacteriophage lambda. J Struct Biol. 2004 Apr-May;146(1-2):72-8. doi: 10.1016/j.jsb.2003.10.021. PMID: 15037238.
Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003 Mar;11(3):659-69. doi: 10.1016/s1097-2765(03)00068-6. PMID: 12667449.
Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002 Dec 27;111(7):1027-39. doi: 10.1016/s0092-8674(02)01198-4. PMID: 12507429.
Herman C, Thévenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 1998 May 1;12(9):1348-55. doi: 10.1101/gad.12.9.1348. PMID: 9573051; PMCID: PMC316767.
Herman C, D’Ari R. Proteolysis and chaperones: the destruction/reconstruction dilemma. Curr Opin Microbiol. 1998 Apr;1(2):204-9. doi: 10.1016/s1369-5274(98)80012-x. PMID: 10066478.
Herman C, Thévenet D, D’Ari R, Bouloc P. The HflB protease of Escherichia coli degrades its inhibitor lambda cIII. J Bacteriol. 1997 Jan;179(2):358-63. doi: 10.1128/jb.179.2.358-363.1997. PMID: 8990286; PMCID: PMC178704.
Herman C, Lecat S, D’Ari R, Bouloc P. Regulation of the heat-shock response depends on divalent metal ions in an hflB mutant of Escherichia coli. Mol Microbiol. 1995 Oct;18(2):247-55. doi: 10.1111/j.1365-2958.1995.mmi_18020247.x. PMID: 8709844.
Herman C, Thévenet D, D’Ari R, Bouloc P. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3516-20. doi: 10.1073/pnas.92.8.3516. PMID: 7724592; PMCID: PMC42198.
Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D’Ari R, Bouloc P. Cell growth and lambda phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10861-5. doi: 10.1073/pnas.90.22.10861. PMID: 8248182; PMCID: PMC47878.








