The advancements of high-throughput transcriptomic, genomic and deep sequencing technologies have generated a flood of data in the public domain and private warehouse. However, the laboratory discoveries based on the analysis of these data have met with limited success. The focus of our laboratory is to apply a multiple disciplinary approach inclusive of bioinformatics, molecular genetics, cancer cell biology, and translational studies to discover driver genetic and epigenetic aberrations, and qualify viable cancer targets on the basis of next-generation sequencing and genomic profiling data.
A. Identification of Oncogene Targets for the Development of Precision Therapeutics in Breast Cancer
Our laboratory research focus is to discover and characterize novel therapeutic targets and predictive biomarkers in breast cancer for the development of precision therapeutics. By interrogating multidimensional genomic datasets with a drug-target database and a “concept signature” analysis we developed to reveal oncogenes (Nature Biotechnology, 2009), we nominated several oncogenes deregulated by genomic amplifications or epimutations as potential therapeutic targets and/or predictive biomarkers in breast cancer. Four candidate targets have been validated by our preliminary experimental and clinical data. We are investigating the clinical relevance and biological function of these targets, as well as their role in breast cancer therapeutic resistance. These projects are funded by the National Cancer Institute (NCI, R01 award), DOD Breast Cancer Idea Award, Nancy Owens Memorial Foundation, DOD Breast Cancer Postdoctoral Fellowship Award, and the Susan G. Komen for the Cure Postdoc Fellowship. We expect that our new discoveries will yield novel insights into recurring genetic and epigenetic abnormalities leading to breast cancer, and establish robust targets for effective and personalized therapies.
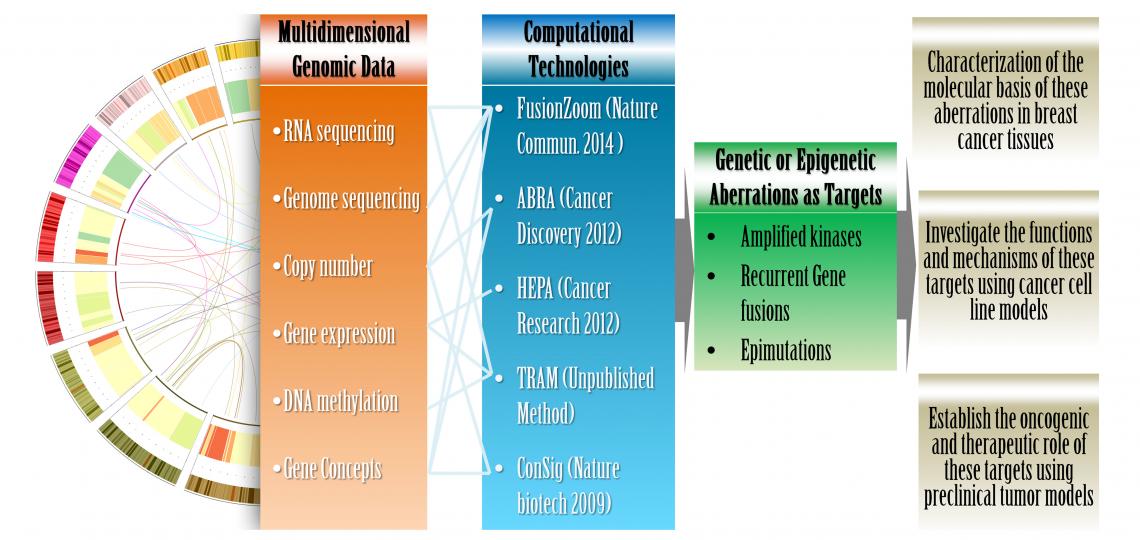
Figure 1. An integrated computational and laboratory approach to translate multidimensional genomic data into viable cancer targets.
B. Characterization of Pathological Recurrent Gene Fusions in Breast Cancer and Other Solid Tumors.
The discovery of TMPRSS2-ETS fusion in ~70% prostate tumors and EML4-ALK in ~7% lung cancer revealed gene fusions as a crucial class of genetic lesions driving epithelial tumorigenesis. To examine the key characteristics that assist in the discovery of recurrent gene fusions in solid tumors, we performed a multi-dimensional characterization of known cancer-related gene fusions. Placing the array of cancer genes in the context of a compilation of “molecular concepts”, including molecular interactions, gene annotations and pathways revealed the “signature concepts” defining the genes driving cancer initiation and progression. Using such information, we developed an innovative concept signature (ConSig) technology that nominates biologically important genetic aberrations from high-throughput data by assessing their association with molecular concepts characteristic of cancer genes.
Special Seminar: A step-by-step guide to Concept Signature (ConSig) Analysis.
To integrate use of high-throughput genomic data, we analyzed the genomic imbalances associated with known gene fusions, finding that recurrent gene fusions exhibit distinctive patterns of copy number alterations corresponding to differential portions of fusion partners. We have formulated this pattern as the “fusion breakpoint principle”, and developed a genome-wide breakpoint mapping analysis to identify recurrent unbalanced rearrangements from copy number data. This principle also laid the foundation for an amplification breakpoint analysis (ABRA) to discover amplified gene fusions in cancer from copy number data (Cancer Discovery 2011).
Figure 2. The Fusion Zoom discovery pipeline: revealing pathological recurrent gene fusions from the scrambled cancer genome.
Based on these principles, we then developed a powerful integrative pipeline called “Fusion Zoom” to reveal recurrent pathological gene fusions from RNA sequencing data (Figure 2). We postulate that the detection of authentic driver gene fusions would be greatly improved by applying more sensitive parameters to comprehensively capture the authentic fusion sequences from the RNAseq data, and by integrating distinct types of genomic data to prioritize the driving fusion events based on the aforementioned principles. The Fusion Zoom pipeline detects recurrent chimeras potentially encoding in-frame protein products from RNAseq data, catalogs the unbalanced breakpoints at the genomic loci of these fusion partner genes from copy number data, and prioritizes pathological gene fusions through the ConSig analysis.
Special Seminar: An introduction the Fusion Zoom pipeline to discover recurrent gene fusions from RNAseq data.
The above analyses have lead to the discovery of recurrent ESR1-CCDC170 fusions in more aggressive breast cancers (Figure 3)(Nature Communications. 2014), recurrent NFE2 rearrangements in lung adenocarcinoma (Nature Biotechnology 2009), and oncogenic KRAS gene fusions in a rare subset of prostate cancer (Cancer Discovery 2012). The ongoing project of our laboratory is to further develop and apply this integrative platform to discover recurrent gene fusions in breast and other major solid tumors. Paired-end RNA sequencing and whole genome sequencing data from the cancer genome atlas (TCGA) will be leveraged to discover chimerical transcripts and genomic rearrangements, and the large measure of cancer genomic data from public domain will be interrogated to facilitate fusion candidate prioritization. This project is funded by The National Cancer Institute (R01 award) and Department Of Defense (postdoc fellowship).
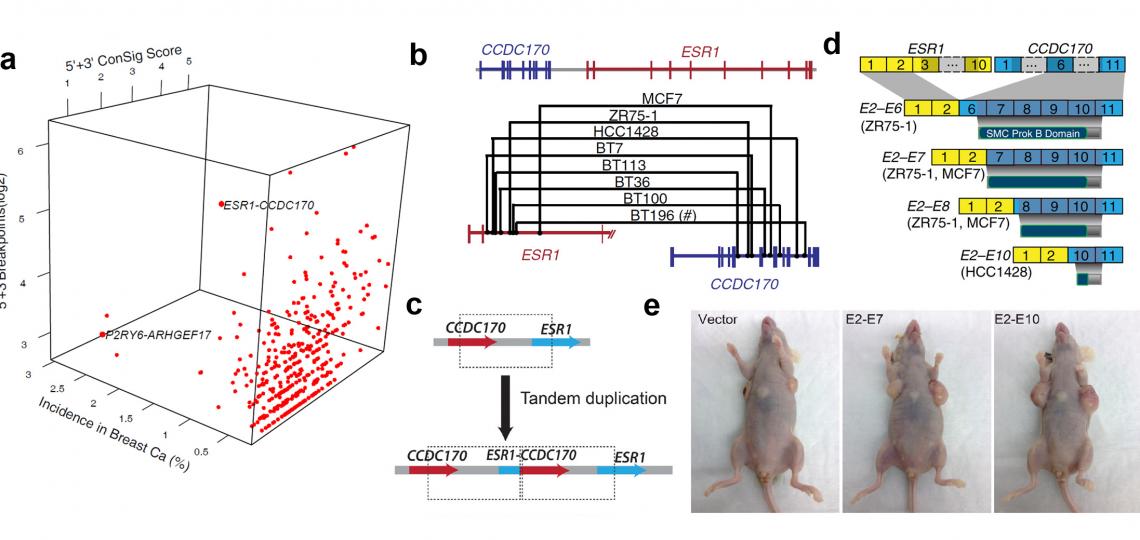
Figure 3. Discover and characterize recurrent ESR1-CCDC170 fusions in an aggressive subset of estrogen-receptor positive breast cancers. (a) Fusion Zoom identifies ESR1-CCDC170 as the lead pathological fusion candidate from RNAseq and copy number data. (b) Genomic fusion points identified by genomic PCR in ESR1-CCDC170 positive tumors. (c) ESR1-CCDC170 may be generated by tandem duplications. (d) ESR1-CCDC170 fusions produce truncated CCDC170 proteins. (e) ESR1-CCDC170 enhanced tumor formation in nude mice when introduced into the breast cancer cells.
C. Genome-Wide Detection of Cancer-Specific Antigen Targets Using an Integrated Computational and Laboratory Technology
Tumor specific antigens (TSAs) have been widely adopted in clinics as active diagnostic and therapeutic targets in cancer. In our previous research project aimed at genome-wide detection of immunological targets, we analyzed the antigens widely adopted as clinical targets, and observed that these antigens usually present a distinctive heterogeneous gene expression profile in large-scale microarray datasets (Figure 4a). We therefore developed the Heterogeneous Expression Profile Analysis (HEPA) which preferentially identifies the clinically useful tumor antigens from the human genome. We then evaluated the immunogenicity of the TSAs by detecting specific autoantibodies in cancer patients. To deal with the large number of candidates, we developed a novel assay called Protein A/G based Reverse Serological Evaluation (PARSE), in which radio-labeled, in vitro translated proteins were used as probes for the presence of serum antibodies (Figure 4b). This allows for quick detection of the autoantibodies against a wide array of serum samples without the need of producing purified recombinant proteins. Further, in this assay, the in vitro translated tumor antigens retained the natural protein conformation and post- translational modifications, thus generating a precise picture of autoantibody responses against these antigens in cancer. Seven out of twelve novel antigens evaluated by PARSE elicited highly tumor-specific autoantibody responses in 4-15% of patients with selected cancers, resulting in distinctive autoantibody signatures in lung and stomach cancers.
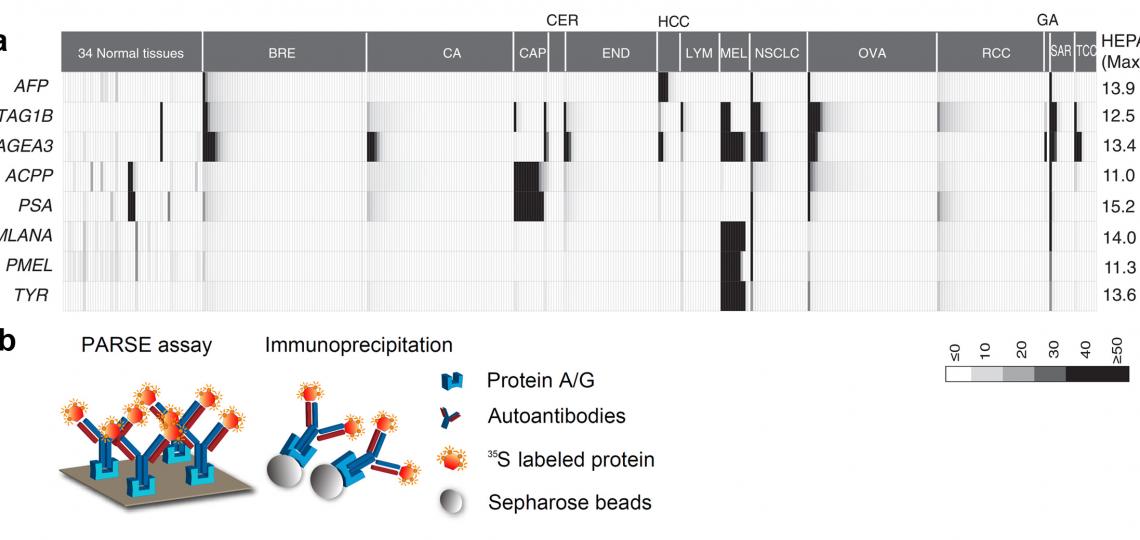
Figure 4. HEPA and PARSE: Systematic discovery of clinically relevant tumor-specific antigens (TSA). (a) The heterogeneous expression profile of clinically important tumor-specific antigens that provided the foundation for heterogeneous expression profile analysis (HEPA). (b) Protein A/Gbased reverse serological evaluation (PARSE) for the quick detection of serum autoantibodies against an array of putative TSA genes.
Together, HEPA-PARSE will comprise an integrative computational-experimental technology for the detection of cancer specific immunome. In addition, the HEPA platform can be also modified and trained to nominate tumor specific membrane targets, which are considered as near-term drug candidates. Combining with the membrane localization database, HEPA can quickly reveal the membrane proteins specific to certain tumor entities. Then the ConSig technology can be applied to evaluate the functional significance of putative membrane targets in cancer progression.
Special Seminar: Heterogeneous expression profile analysis (HEPA): genome-wide discovery of tumor specific antigen targets in cancer.








 Credit
Credit
