Our software, tools, datasets, etc. are open-source, and free for anyone around the world to use and modify. We strive to make resources that are high quality in every aspect: cleanly written, robustly constructed and tested, well-documented, easy-to-use, accessible, customizable, and as effective as possible in real-world use.
Genetics
Gene Ranking, Identification and Prediction Tool
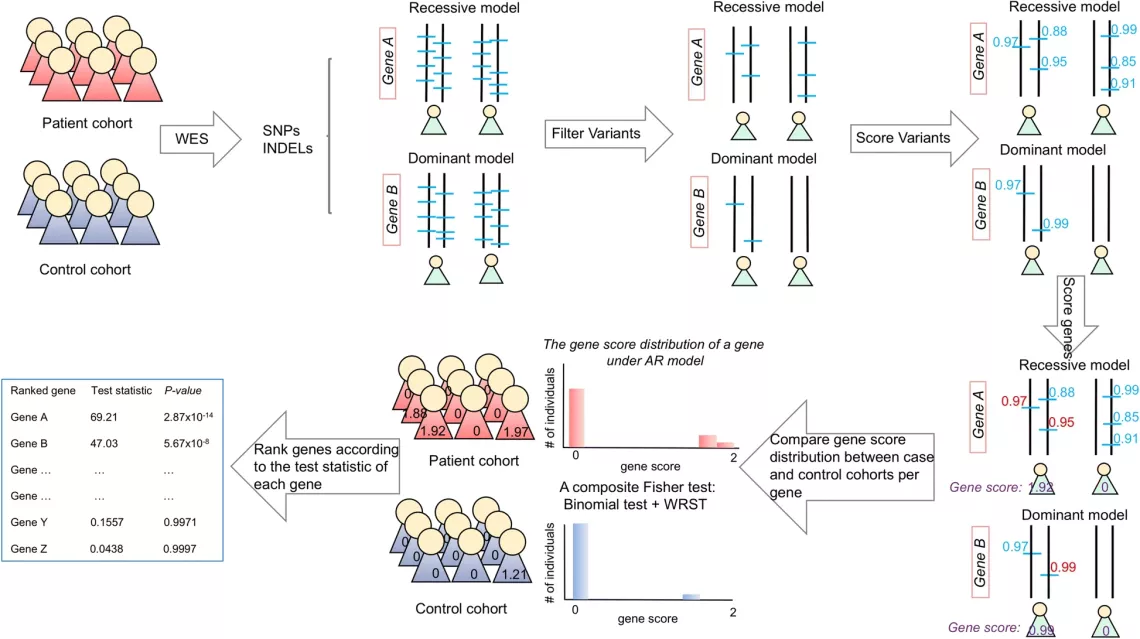
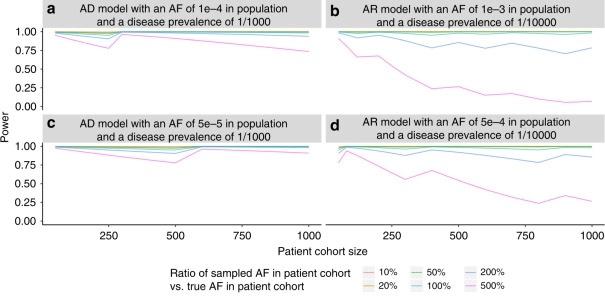
A novel method, the Gene Ranking, Identification and Prediction Tool (GRIPT), for performing case-control analysis of NGS data which is well-powered for disease gene discovery, especially for diseases with high locus heterogeneity.
Macaque Genome Database
The Macaque Genome Database is a coalition of investigators seeking to aggregate and harmonize exome sequencing data from a wide variety of large-scale sequencing projects of Macaque. You need to access under BCM network.
Binomial Test
A novel statistical test by combining sequencing data from a patient cohort with a normal control population database. By comparing the expected and observed allele frequency in the patient cohort, variants that are likely benign can be identified.
Single Cell
Single-cell Transcriptomic Atlas for Adult Human Retina
A fractionation protocol was developed to enrich nuclei from rare neuron cell types, including bipolar cells, amacrine cells, and retinal ganglion cells. In total, over 60 cell types are identified in our dataset, making the currently most comprehensive single-cell profiling of adult human retina.
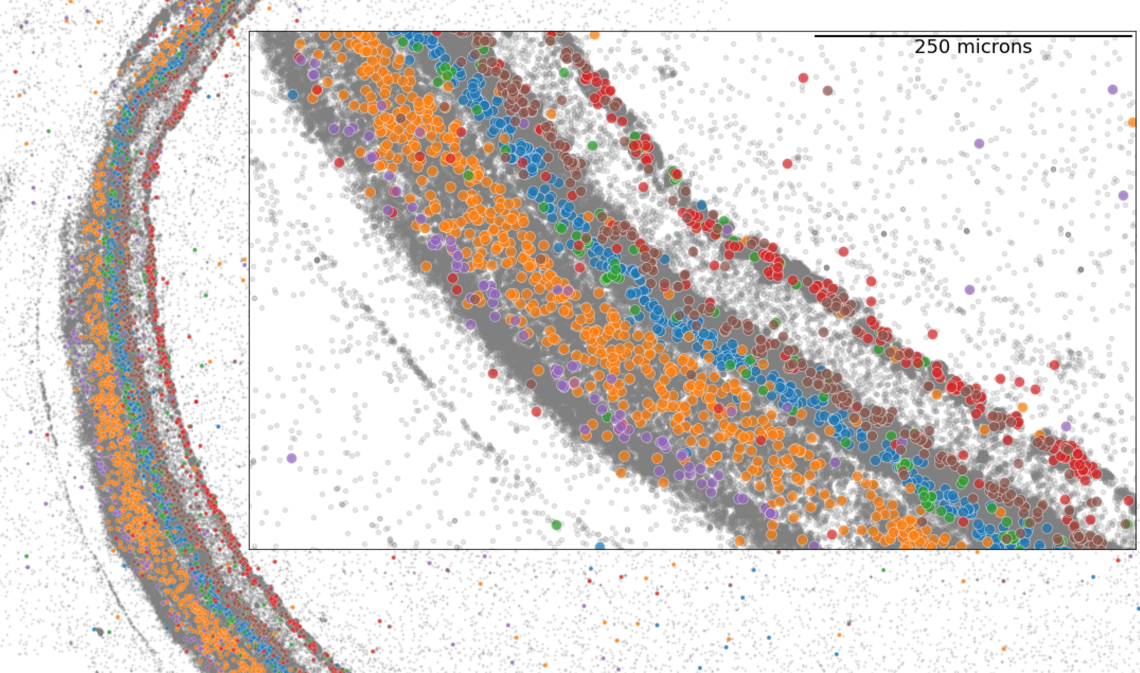
Spatial Atlas of Mouse Retina
Spatial atlas of mouse retina in MERFISH experiment. The web service visualizes the spatial organization of mouse retina through cellxgene. Imputed data enable gene expression exploration of the whole transcriptome.
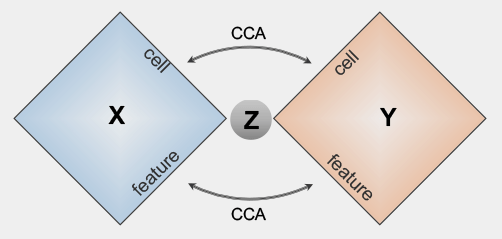
bindSC
bindSC is an R package for single cell multi-omic integration analysis. It is developed to address the challenge of single-cell multi-omic data integration that consists of unpaired cells measured with unmatched features across modalities.
Resource Links
Gulf Coast Cluster for Single Cell Omics
The GCC single cell omics cluster aims at leveraging the expertise at different institutions and maximize the interdisciplinary synergy of the group.
Eye Biological Network
The Eye Biological Network has established a comprehensive molecular and spatial atlas of the cells in the visual system across age and ethnicity groups. This resource serves the foundation for further dissection of the function, interaction, and involvement of subtypes of cells in the visual function and diseases.
Network Coordinators: Rui Chen, Ph.D. Email
Single Cell Genomics Core
The Single Cell Genomics Core provides comprehensive services for RNA and chromatin profiling on single or a small number of cells. The core is equipped with the 10x Genomics Chromium Platform, the Takara icell8 platform, and the Fluidigm C1 auto cell prep platforms. The combination of these platforms allows cost-effective and high throughput genomic profiling that matches with the experiments.
Academic Director: Rui Chen, Ph.D. Email
X-linked Inherited Retinal Disease Variant Curation Expert Panel
We will have established variant curation rules that can be applied for all X- linked IRD genes, completed the curation of these IRD genes, and established a framework that can be modified and applied to other genes affecting inherited retinal disease.
Genetics
References:
Effect Predictor & others:
Single Cell
Tools: