Our laboratory has developed a range of data analysis workflows that incorporate advanced statistical and computational methods to interpret the complex molecular datasets generated by MS technologies. A major focus of our work is the application of logistic regression and machine learning approaches, particularly the Least Absolute Shrinkage and Selection Operator (Lasso) to build robust statistical classifiers and identify predictive molecular features. The “Lasso” technique offers significant advantages in high dimensional omics datasets by performing simultaneous variable selection and regularization. This enables the identification of molecular features most strongly associated with disease states while constructing sparse, interpretable models capable of accurately predicting cancer and supporting tissue diagnosis.
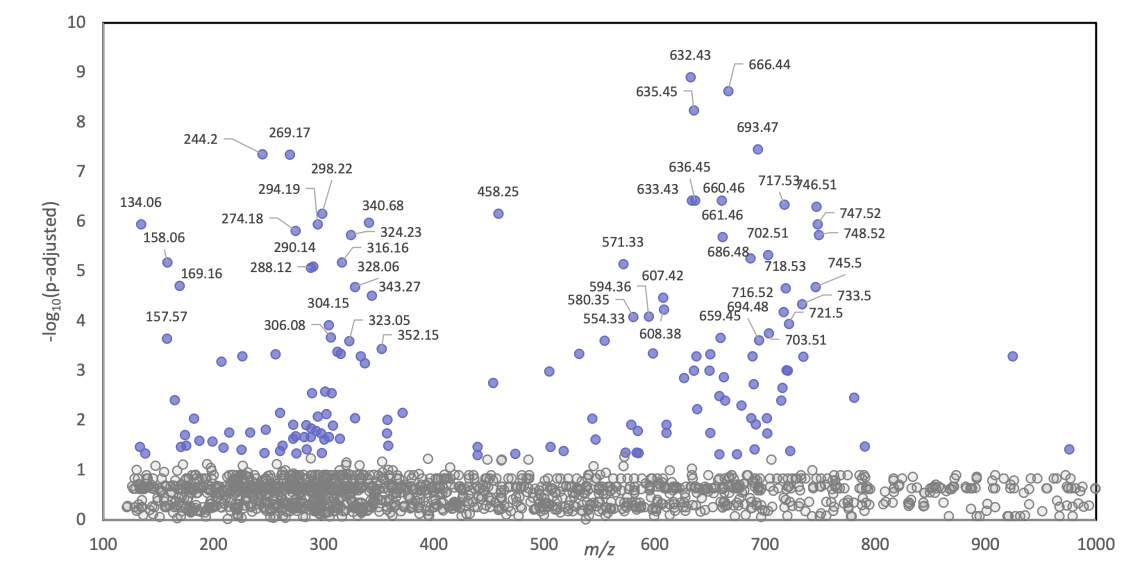
In addition, we have adapted the Significance Analysis of Microarrays (SAM) framework for use with MS imaging datasets. “SAM” allows us to systematically identify molecular markers that differ significantly across biological conditions while controlling for false discovery rates. This approach has been widely used in the scientific community to analyze high dimensional imaging and omics data, and we continue to refine these methods to further enhance rigor, reproducibility, and robustness in our analyses.
We are currently developing new methods for MasPec data visualization and processing, and all tools are still in the testing phase.
All these efforts aim to enhance the resolution, integration, and interpretability of spatial multi omics datasets, ultimately enabling deeper molecular insights into human disease, supporting the discovery of novel therapeutic targets and improving the clinical and biological insights derived from MS datasets.








