Our laboratory uses genetic, cell biological, and biochemical approaches to explore the pathogenesis of polyglutamine neurodegenerative diseases, the function of Math1 in neurodevelopment, and how MECP2 mutations cause postnatal neurodevelopmental disorders.
Polyglutamine Neurodegenerative Disorders
The spinocerebellar ataxias (SCAs) are dominantly inherited neurodegenerative disorders characterized by a progressive loss of balance and coordination, and eventually the inability to coordinate swallowing and breathing. In collaboration with Harry Orr's group (University of Minnesota), we determined that the mutation responsible for SCA1 is an expansion of a CAG trinucleotide repeat encoding glutamine in the protein Ataxin-1. Our efforts have focused on addressing how increasing the number of glutamines in Ataxin 1 causes neuronal degeneration.
Genetic studies in mice and fruit flies have shed the most light on SCA1 pathogenesis. Glutamine-expanded Ataxin-1 causes disease by a gain-of-function mechanism given that mice lacking this protein do not reproduce SCA1 features. Our analysis of SCA1 transgenic mice revealed that the mutant protein aggregates in the nuclei of cerebellar Purkinje cells and that these aggregates stain positively for ubiquitin, the proteasome, and some chaperones. This led us to propose that protein misfolding or impaired protein clearance might be critical in SCA1 pathogenesis. We tested this hypothesis in cells, fruit flies, and mice and found that indeed chaperone overexpression mitigates SCA1 neuropathology and phenotypes. We created a mouse model that expresses the glutamine-expanded Ataxin-1 under the control of endogenous locus (knock-in SCA1 mice) and found that this model recapitulates all features of SCA1. Importantly, from this model we learned that neurons that develop nuclear inclusions last are the most vulnerable. This suggested to us that the nuclear inclusions are probably protective by sequestering the glutamine-expanded protein and reducing its native interactions.
In collaboration with Juan Botas (Baylor College of Medicine), we found that high levels of wild-type Ataxin-1 produce effects similar to mutant Ataxin-1 in Drosophila and mice. This led us to propose that wild-type Ataxin-1 might take on a conformation that resists clearance or interacts strongly with other proteins and that such conformation is favored by the expanded polyglutamine tract. This model predicts that most proteins that interact with wild-type Ataxin-1 should interact with the mutant protein and that modifications of Ataxin-1 that might alter its conformation are critical to pathogenesis. This proved to be the case. In collaboration with the Orr lab, we found that residue S776 in Ataxin-1 is necessary for Ataxin-1's toxicity. We found that 14-3-3 proteins interact with Ataxin-1 in a S776-dependent manner and that 14-3-3 augments levels of Ataxin-1 when phosphorylated at S776 by Akt kinase. Second, we found that the AXH domain in Ataxin-1 is a key mediator of the neuropathology in part through interactions with the transcriptional repressors Gfi-1 and Capicua. The glutamine tract seems to mediate its toxicity by modulating the activity of the AXH domain. Third, we found that mutant Ataxin-1 has to be in its native complexes (that contain Capicua) to make its pathogenic affects. Lastly, we recently found that the Ataxin-1 paralog (Ataxin-1 like) can suppress neurodegeneration in the SCA1 knock-in mice by displacing Ataxin-1 from its native complex facilitating its sequestration into inclusions.
Recognizing the importance of protein interactions for mediating SCA1 pathogenesis, and given that SCA1 shares clinical and pathological features with several other inherited ataxias, we proposed that understanding the interactions of Ataxin-1 as they relate to the interactions of proteins implicated in other ataxias might provide better insight about molecular mechanisms leading to Purkinje cell degeneration and ataxia. To test this hypothesis we used proteins whose genes are mutated in various ataxias to develop a "phenotype-based" protein interaction network (ataxia interactome) in collaboration with Marc Vidal at the Dana-Farber Cancer Institute. To our surprise we found that several ataxia proteins interact directly or indirectly and that certain RNA-binding proteins and transcription regulators form key hubs in the ataxia network. Moreover, several of the direct physical interactors are modifiers of disease phenotypes in animal models. We are currently investigating several protein interactions within the network to gain better insight about SCA1 pathogenesis and the pathogenesis of other poly-glutamine diseases such as SCA6 and SCA7. In addition we are performing genetic and biochemical studies to gain better insight about the in vivo function of the native Ataxin-1 complexes and the role of Capicua in the nervous system.
Math1 and Neurodevelopment
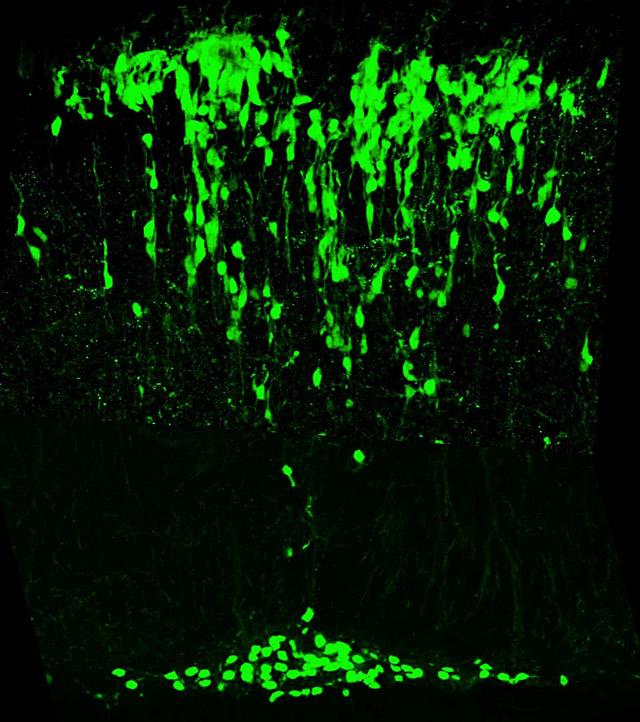
We isolated the mouse atonal homologue 1 (Math1) because of the importance of atonal for the development of balance and hearing organs in Drosophila.
Mice lacking Math1 die at birth because they cannot initiate respiration. These mice lack cerebellar granule neurons, pontine neurons, hair cells in the vestibular and auditory systems, and the D1 interneurons of the spinocerebellar tracts. That a single gene controls the genesis and/or differentiation of multiple components of the proprioceptive pathway was a surprise. Math1 is also essential for secretory cells in the gut (Paneth, goblet, and enteroendocrine cells), and the enteroendocrine cells secrete neuropeptides that modulate gut proprioception. Recently we discovered that Math1 also controls multiple components of the auditory and vestibular pathways, and within the cerebellum it controls the genesis of some deep cerebellar neurons in addition to granule neurons. Identification of the Math1-dependent neurons allowed us to propose that Math1 redefines the rhombic lip and its derivatives.
We are continuing to characterize the Math1-null mice to pinpoint the neurons involved in the respiration phenotype, and we are pursuing the identification of Math1's downstream targets to define the molecular pathways involved in the differentiation of the diverse cell types dependent on Math1.
Rett Syndrome and MeCP2
Girls affected with Rett syndrome appear to develop normally for the first 6 to 18 months of life, then lose the ability to speak and socialize, and develop tremors, ataxia, seizures, and stereotypic hand-wringing movements. In 1999, we discovered that Rett syndrome is caused by mutations in the gene encoding methyl-CpG-binding protein 2 (MECP2). Located on the X chromosome, MECP2 encodes a protein that binds methylated cytosines, helping to orchestrate gene silencing via DNA methylation.
We and others have discovered that MECP2 mutations cause a broad spectrum of phenotypes in both females and males. Females may present with isolated mental retardation, autism, or milder forms of Rett if they have favorable X-chromosome inactivation. In males, the inactivating mutations cause severe neonatal encephalopathy and death in infancy, whereas milder mutations may cause mental retardation, motor dysfunction, and psychosis.
Our analysis of the cellular distribution of MeCP2 during development determined that MeCP2 is in mature neurons and that the number of MeCP2-positive cortical neurons increases up to 10 years of age in humans. We generated a mouse model by creating a mutation that truncates the protein post–amino acid 308. Male Mecp2308 mice appear normal up to six weeks of age, when they develop tremors, seizures, coordination problems, social behavior abnormalities, and forepaw stereotypies similar to the hand-wringing seen in patients. We also generated transgenic mice that overexpress MeCP2 at twice the normal level in the correct spatiotemporal distribution, and found that they develop a progressive postnatal neurodevelopmental disorder. This led us to propose that duplications of MeCP2 might lead to postnatal neurologic disorders, which indeed is proving to be the case. We have begun to characterize patients with the duplication and found that some have Rett-like features, while others have autism spectrum phenotypes.
Our functional and pathogenesis studies, revealed that MeCP2 interacts with the RNA-binding protein YB-1 and that this interaction affects alternative splicing of reporter cassettes regulated by YB-1. The finding that Mecp2308 mice have altered RNA-splicing patterns in the cerebral cortex that are significantly different from these of wild-type animals led us to propose that MeCP2 acts as repressor, but when it is released from the promoter region, it participates in its second function as a splicing regulator of the newly transcribed genes. We are studying both the Mecp2308 and overexpression mice to identify the neuron-specific expression and splicing changes that result from MeCP2 dysfunction. We are collaborating with Nathaniel Heintz at Rockefeller University to isolate cell-specific RNAs using the BACarray technology developed in his lab. Recently we found that MeCP2 regulates the expression of corticotropin-releasing hormone (CRH) by binding the promoter of the Crh gene. The elevation of the Crh level in the Mecp2308 mice could explain the anxiety-like phenotype and the exaggerated poststress corticosterone response in these animals. We are treating Mecp2308 mice with Crh receptor-1 antagonist to determine if this will subdue the anxiety and stress phenotypes. To determine the anatomical substrates for the various Rett phenotypes we are generating mice that lack MeCP2 in specific neurons. Our ultimate goal is to correlate phenotypes to specific neurons and to identify the gene expression changes within such neurons that could be modulated to modify the phenotypes.








