
2011 Lab Photo
From left: Mohan Joshi, David Magnan, Nick Stepankiw, Mark Lies, Shana Williams, David Bates

Chromosome Painting
Four-color in situ hybridization paints in E. coli.
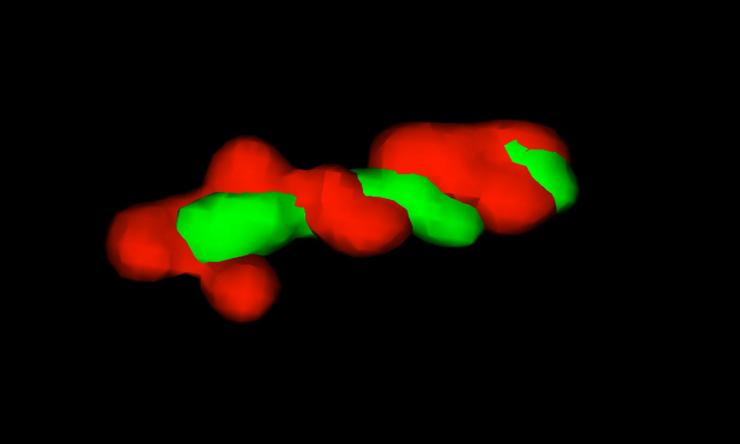
G1 Chromosome Arms
3D projection of left and right chromosome arms.
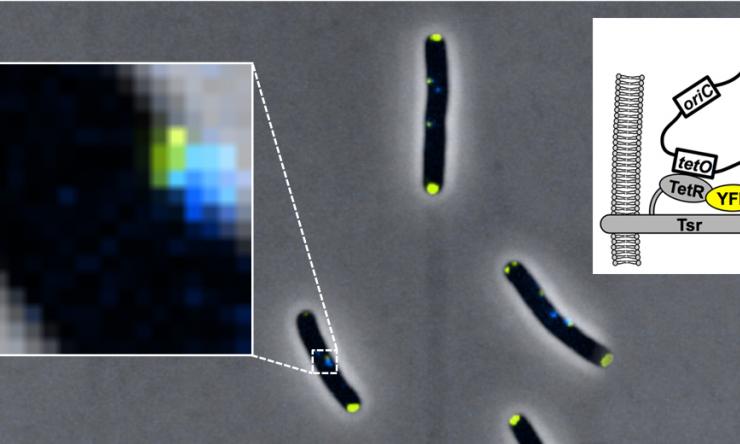
Origin Tethering
Origin tethering by Tsr-TetR-YFP fusion protein.
Research Areas
Chromosome Dynamics
Although genetic information is encoded in a one-dimensional array of DNA bases, all major DNA processes (replication, transcription, and recombination) are controlled by changes in the three-dimensional structure of DNA. Large-scale structural features of chromosomes including the arrangement of important chromosomal sites (origins, termini, and centromeres) and overall chromosome compactness change dramatically and predictably during the cell cycle. One major limitation in studying chromosome dynamics is our inability to observe large-scale chromosome structure using standard fluorescence microscopy methods, which only label a discrete genetic position (focus). We are developing a novel chromosome painting technology to image individual domains within the entire chromosome in single cells. This method, inspired by in situ hybridization-based human karyotyping techniques, utilizes multi-color combinatorial labeling and high-resolution three-dimensional photography to generate whole genome maps of the chromosome. Our goal is to define the normal program of chromosome movement in E. coli using a cell cycle synchronization apparatus we designed called the “baby cell machine” (Bates et al, 2005).
Chromosome Cohesion
In eukaryotic DNA replication, sister chromosomes are attached to each other (cohesion) until they are separated at nuclear division. By aligning sister chromosomes side-by-side along homologous sequences, chromosome cohesion enables precise attachment and segregation by the mitotic spindle apparatus and aids efficient repair of DNA damage by homologous recombination. Eukaryotic chromosome cohesion involves both a proteinaceous component, the cohesin complex, and physical entanglement of sister chromosomes, but the relative roles of these components in maintaining and regulating cohesion is unknown. We and others have shown that newly replicated DNA in E. coli incurs a transient cohesion period, with most regions remaining attached for 7-10 minutes before separating apart to opposite cell poles (Nielsen et al, 2006; Espeli et al, 2008; Wang et al, 2008; Joshi et al, 2011). Thus, forks are trailed by a 300-400 kb sliding window of sister chromatid cohesion from origin to terminus. We later showed that cohesion could be directly modulated by changing the cellular abundance of a type II topoisomerase called Topo IV, a homolog of Human TopoII (Joshi et al, 2013). As Topo IV is the primary enzyme for resolution of entanglements between sister chromosomes, our data suggest that cohesion likely results from precatenation of nascent duplexes as they exit the fork (see figure). Our evidence suggests that decatenation by Topo IV is delayed by a DNA binding protein called SeqA, which binds to hemimethylated GATC sites behind the fork until newly replicated GATC's are remethylated by Dam methylase 5-10 minutes later. We hypothesize that SeqA inhibits Topo IV by changing DNA curvature, which is needed for Topo IV recognition of catenated segments (Bates et al, 2016). Interestingly, cohesion lasts much longer at a cluster of sites (termed "Snaps") on the right chromosome arm, which separate in unison midway through the replication cycle promoting efficient chromosome segregation (Joshi et al, 2013).
DNA Topology at the Replication Fork
The presence of extensive DNA catenation behind the replication fork implies that DNA at the fork is under extreme helical tension in the form of positive supercoiling, that exceeds the relaxing ability of topoisomerases (forks can travel up to an astounding 1000 bp/sec!). In theory, precatenanes are formed when positive supercoils migrate behind the fork via rotation of the fork structure, including the ~800 kDa replisome and nascent sister duplexes. Our lab is currently investigating how this tension affects fork progression, which likely ranges from fork slowing to complete fork arrest, and what mechanisms allow forks to replicate past topological barriers (Sneak Peak). We are also measuring supercoiling across the genome to determine which factors lead to topological strain, including topological insulators like DNA bound proteins and transcription complexes, and which factors alleviate topological strain at the fork, including topoisomerases, accessory helicases, replication factories (association between leftward and rightward replisomes), and cohesion.
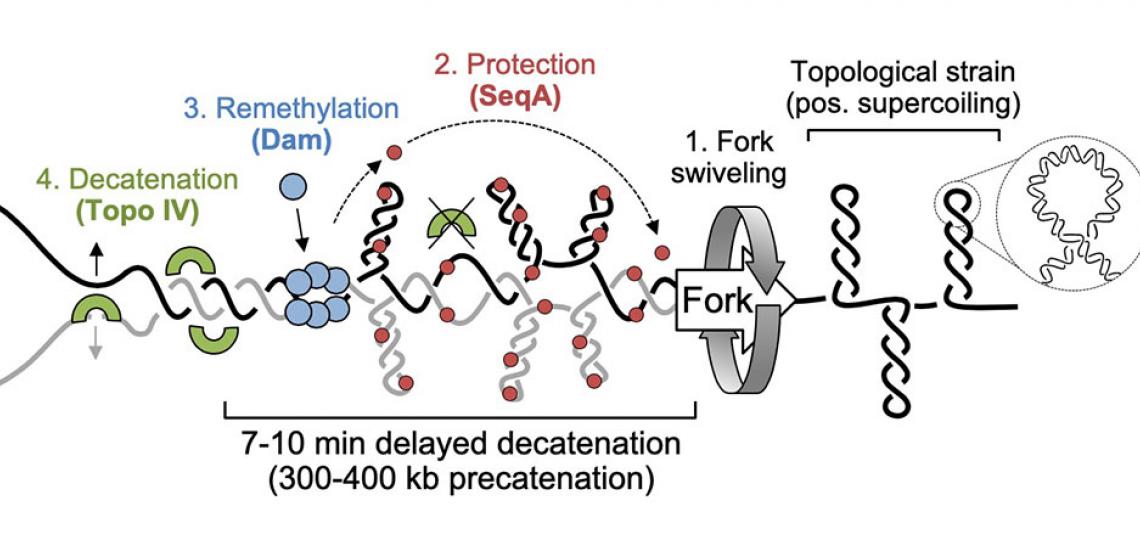
Precatenane formation and resolution at the fork
Our model for precatenane formation and resolution in E. coli (adapted from Joshi et al, 2013). Topological strain ahead of the replication fork causes fork swiveling and catenation of sister chromatids. SeqA protein binds newly replicated (hemimethylated) DNA and inhibits Topo IV-mediated precatenane resolution, possibly by altering the geometry of inter-sister links. After 7-10 minutes, Dam remethylates newly synthesized strands, releasing SeqA and allowing Topo IV to resolve sister chromatids.








