A majority of proteins must fold correctly in order to attain biological function. Although the protein folding information is encoded within the nascent polypeptide chain, newly synthesized polypeptides and those imported into organelles are prone to misfolding, causing aggregation and formation of other toxic species. Consequently, maintaining protein homeostasis (proteostasis) is essential for cell and organismal health. To accomplish this, cells have evolved a sophisticated network of protein quality control machines consisting of molecular chaperones and energy-dependent proteases, which monitor the protein folding and their assembly into functional complexes, and selectively remove excess and damaged proteins from the cell.
Molecular chaperones are a group of proteins that assist other macromolecules in the folding and assembly of higher order structures without being components of these final structures. It is now known that molecular chaperones not only promote protein folding in the “forward” direction by facilitating folding and preventing misfolding and aggregation, but also facilitate protein unfolding and even disaggregation by recovering functional protein from aggregates (Jeng et al. F1000Res. 2015). A research focus of the Tsai laboratory is to understand how ATP-dependent molecular chaperones, such as Hsp60 (GroEL), Hsp70 (DnaK), Hsp90 (HtpG), and Hsp100 (ClpB), harness metabolic energy to facilitate protein folding. Hsp100 chaperones are unusual in that they do not promote protein folding but instead cooperate with the cognate Hsp70 system (Hsp70/Hsp40/NEF) to recover functional proteins from aggregates, an activity that is termed protein disaggregation.
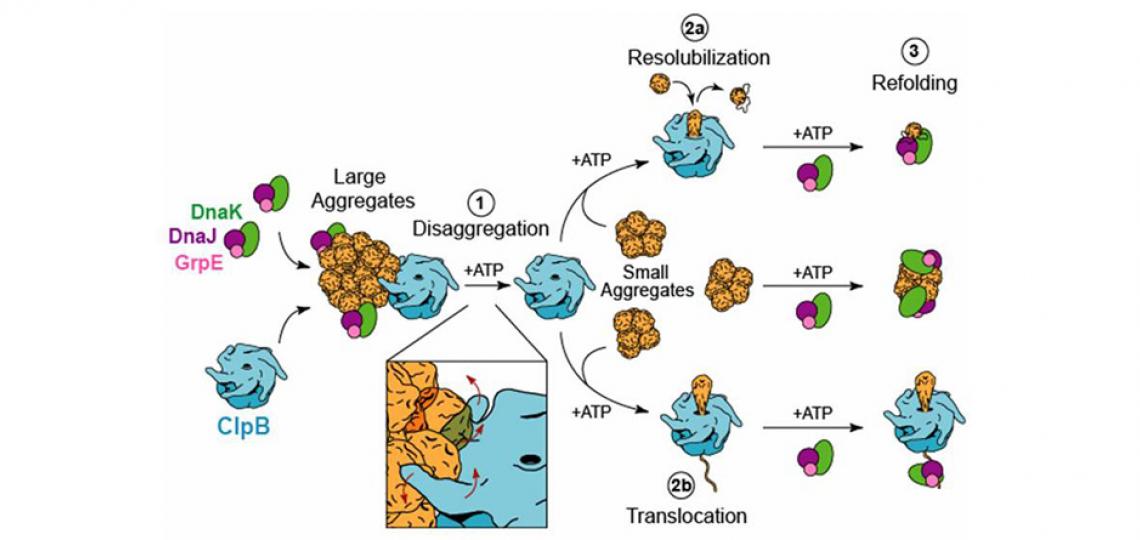
Figure 1: Hsp100 chaperones facilitate the disaggregation of amorphous protein aggregates and amyloid fibers (ordered aggregates) (Lee et al. Cell 2003)
To investigate the structure-function relationship of ClpB, we have engineered a ClpB variant, known as BAP that, unlike ClpB, associates with the ClpP protease in an ATP-dependent manner. While ClpB and BAP share the ability to disaggregate proteins, BAP (but not ClpB) functions as a novel disaggregating-degrading machine in the presence of ClpP. Using BAP, we demonstrated that substrates must translocate through the central pore of the ClpB hexamer and, perhaps more importantly, that thermotolerance requires the refolding of aggregated proteins, i.e. it is not the aggregate itself that causes cell death (Weibezahn et al. Cell 2004).
While the structure-function relationship is beginning to be understood, the ATP-driven conformational changes that occur in the ClpB hexamer are unknown. Moreover, it remains unclear how the nucleotide-driven conformational changes are coupled to substrate recognition and binding. To provide the structural basis for substrate binding, we determined the single-particle, cryo-EM reconstruction of the ATP-activated state of ClpB (EMDB: 1244) by examining the structure of a ClpB mutant that binds but does not hydrolyze ATP (Trap-ATP). In addition, we also obtained single-particle reconstructions of ClpB wild-type in the AMPPNP (EMDB: 1243), ADP (EMDB: 1242), and nucleotide-free states (EMDB: 1241). Each structure represents a snapshot of ClpB in a different nucleotide-state, and together the structures provide a molecular understanding of the ATP-driven conformational changes as they may occur in solution (Lee et al. Mol. Cell 2007).
More recently, we have focused on Hsp104 the yeast ortholog of ClpB. In addition to its role in induced thermotolerance development, Hsp104 is also required for the inheritance and maintenance of all amyloid-forming yeast prions. Surprisingly, Hsp104 was proposed to be an atypical AAA+ machine that differs in three-dimensional structure from ClpB. To address this issue, we determined the cryoEM structures of an Hsp104 Trap variant in the ATP-bound state (EMDB: 1631) and the structure of an Hsp104-T4 Lysozyme chimera (EMDB: 1630), which has gained the ability to rescue heat-aggregated proteins in the absence of Hsp70/40 chaperones (Lee et al. PNAS 2010). Our work provides direct evidence that Hsp104 is not an atypical AAA+ machine as previously proposed, but resembles ClpB and other AAA+ machines in three-dimensional structure. To understand the role of the M-domain in protein disaggregation, we generated Hsp104-ClpB chimeras that feature the M-domain from the orthologous protein and demonstrated for the first time that the M-domain directly controls the ClpB/Hsp104 protein remodeling activities in a species-specific and Hsp70-dependent manner (Sielaff and Tsai, JMB 2010) and is required for Hsp70 binding that activates the Hsp104 motor (Lee et al. PNAS 2013).








