Chemical approach created to discover compounds for drug development
Scientists at Baylor College of Medicine have devised a chemical approach for discovering new enzyme-binding compounds that could be developed into drugs for various illnesses and conditions. This process can be used to support development of novel antiviral drugs for use in the continuing COVID-19 pandemic.
The study, published online today in the Proceedings of the National Academy of Sciences USA, was co-led by Dr. Surendra Dawadi, instructor in the Center for Drug Discovery at Baylor, and Dr. Martin M. Matzuk, professor and interim chair of the Department of Pathology & Immunology and director of the Center for Drug Discovery at Baylor.
“The study demonstrates that a powerful drug discovery approach, DNA-encoded chemical library technology, can be successfully used for the discovery of drug-like small molecules to inhibit some classes of protease enzymes, which are involved in various pathophysiological processes related to disease or injury,” said Dawadi.
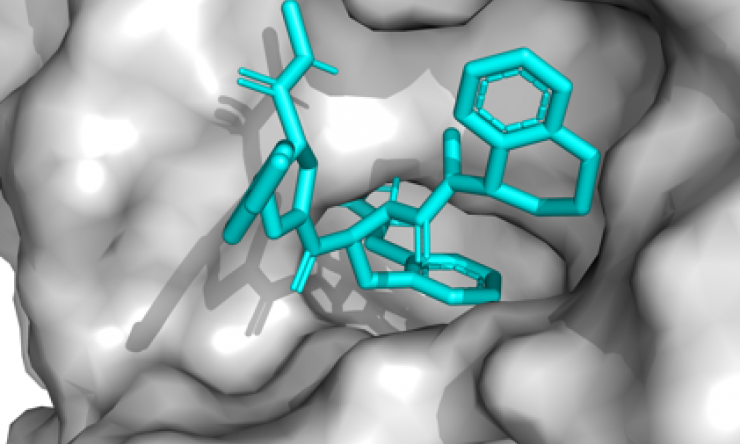
Although DNA-encoded chemical library technology for drug discovery has existed for more than 20 years, Baylor researchers developed new chemical methods to synthesize structurally focused chemical libraries designed to possess high affinity for a particular class of protein targets, in this case proteases. Proteases, enzymes that catalyze the hydrolysis of peptidic bonds in proteins, have been an intensely studied class of targets for drug discovery due to their involvement in various pathophysiological processes.
To rapidly identify small-molecule lead compounds to target healthcare-associated proteases, the researchers constructed a 9.8 million membered protease-focused DNA-encoded chemical library, the first example of this type of library. To evaluate this library, the team focused on thrombin, a common protease, and found an inhibitor of thrombin that is 10-fold more potent than some clinical compounds.
“This finding means that our collaborative team of medicinal chemists, biochemists and computational scientists can use this focused library approach to identify potential drug targets for other proteases, including those that play essential roles in infectious diseases,” Matzuk said. “Because a few proteases have been shown to be relevant to the pathogenesis of COVID-19, similar screens of this and currently developed protease-focused DNA-encoded chemical libraries are critical and ongoing to develop novel antiviral drugs.”
These findings also have implications for health issues where no drugs exist, such as male contraception.
Other Baylor College of Medicine co-authors on this paper are Nicholas Simmons, Gabriela Miklossy, Kurt M. Bohren, John C. Faver, Melek Nihan Ucisik, Pranavanand Nyshadham, and Zhifeng Yu. Funding for the study is provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the Bill and Melinda Gates Foundation, the Welch Foundation, a Core Facility Support Award from the Cancer Prevention Research Institute of Texas, and a COVID-19-relevant exploratory grant from Baylor College of Medicine.







 Credit
Credit